All content on this site is intended for healthcare professionals only. By acknowledging this message and accessing the information on this website you are confirming that you are a Healthcare Professional. If you are a patient or carer, please visit Know AML.
The aml Hub website uses a third-party service provided by Google that dynamically translates web content. Translations are machine generated, so may not be an exact or complete translation, and the aml Hub cannot guarantee the accuracy of translated content. The aml and its employees will not be liable for any direct, indirect, or consequential damages (even if foreseeable) resulting from use of the Google Translate feature. For further support with Google Translate, visit Google Translate Help.
The AML Hub is an independent medical education platform, sponsored by Daiichi Sankyo, Johnson & Johnson, Syndax, Thermo Fisher Scientific, Kura Oncology, AbbVie, and has been supported through an educational grant from the Hippocrate Conference Institute, an association of the Servier Group.
Funders are allowed no direct influence on our content. The levels of sponsorship listed are reflective of the amount of funding given. View funders.
Now you can support HCPs in making informed decisions for their patients
Your contribution helps us continuously deliver expertly curated content to HCPs worldwide. You will also have the opportunity to make a content suggestion for consideration and receive updates on the impact contributions are making to our content.
Find out more
Create an account and access these new features:
Bookmark content to read later
Select your specific areas of interest
View AML content recommended for you
2022 ELN recommendations for the diagnosis of AML in adults
Do you know... Which of the following genetic abnormalities is classified within the adverse-risk category in the 2022 ELN recommendations?
The European LeukemiaNet (ELN) has previously published guidelines for the diagnosis and management of acute myeloid leukemia (AML) in 2010 and 2017.1 Due to the major advances in the classification of AML, genomic diagnostics, and molecular markers of disease, the ELN has recently released a 2022 update, which includes revised genetic risk classification.1 Döhner, et al.1 published these recommendations on behalf of the ELN in Blood, and we are pleased to summarize these latest recommendations here.
Methods
An international panel of clinical and research experts carried out comprehensive literature searches of the PubMed, Cochrane, and Medline databases for publications from 2017 onwards using the National Comprehensive Cancer Network (NCCN) categories of evidence.1,2
Disease classification
Currently, genetic abnormalities define the classification of AML disease within the International Consensus Classification of AML, with additional features as qualifiers of the primary diagnosis (Figure 1).
Figure 1. Hierarchical classification of the International Consensus Classification of AML*
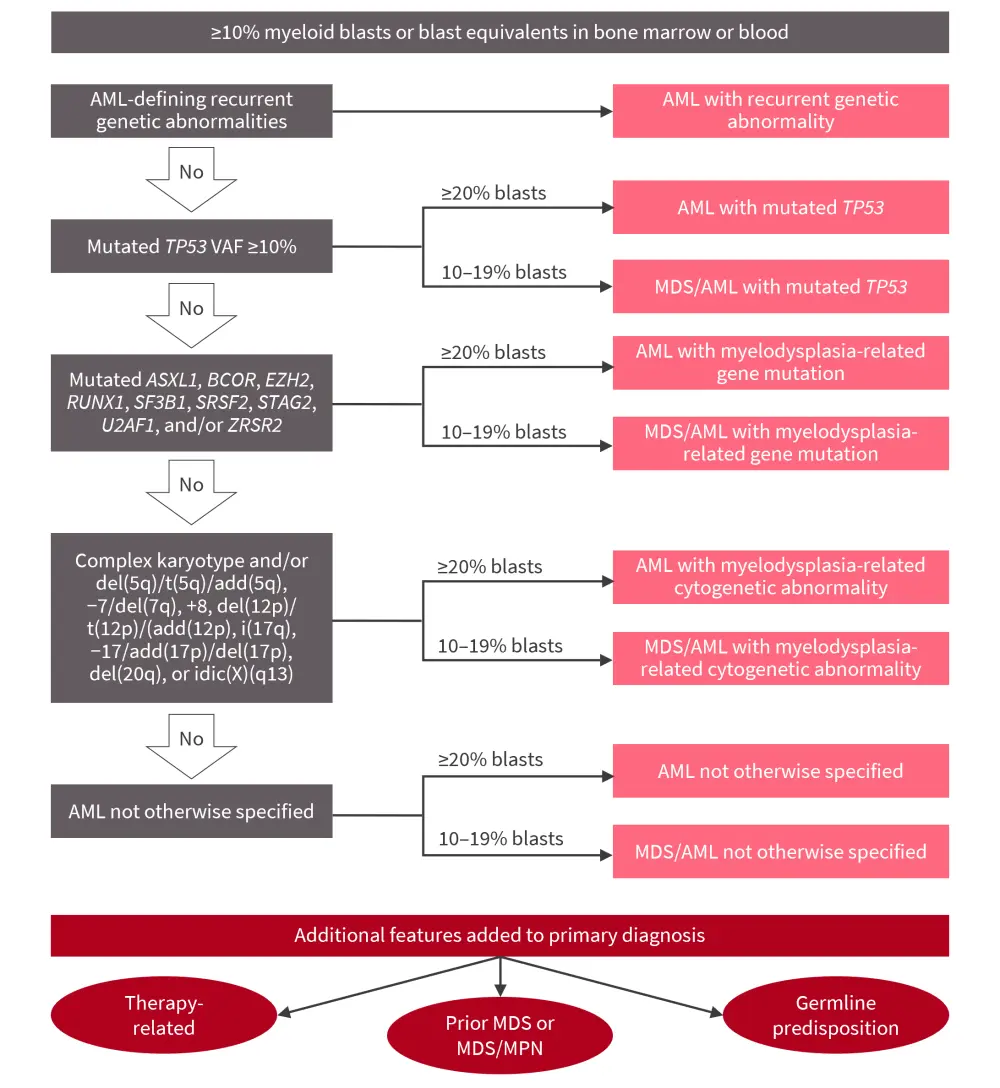
AML, acute myeloid leukemia; MDS, myelodysplastic syndromes; MPN, myeloproliferative neoplasms; VAF, variant allele frequency.
*Adapted from Döhner, et al.1
While the blast threshold remains at 20% for the majority of AML subtypes, the recurrent genetic lesions, shown in Figure 2, are now considered to indicate AML when there are ≥10% bone marrow (BM) or blood blasts. This allows patients with low-blast count AML to access treatment approaches for myelodysplastic syndromes (MDS) and AML. It also allows patients with low-blast count AML to be eligible for both AML and MDS clinical trials, providing them with a wider range of available therapies, as covered previously on the AML Hub. Therefore, the following categories (also shown in Figure 1) are classified as AML if there are 20% BM or blood blasts or as AML/MDS if the BM or blood blast count is 10–19%.
- AML with recurrent genetic abnormalities (Figure 2)
- An expansion of the previous category to include additional genetic translocations
- Any in-frame bZIP CEBPA mutation, rather than the previous requirement of biallelic CEBPA abnormalities
- AML with mutated TP53
- TP53-mutated AML and MDS with a variant allele frequency >10% should be considered a separate disease entity; TP53 mutations are associated with a very poor prognosis, occurring in the absence of other AML-associated mutations in around half of cases
- AML with myelodysplasia-related gene mutations
- Mutations in ASXL1, BCOR, EZH2, RUNX1, SF3B1, SRSF2, STAG2, U2AF1, and/or ZRSR2 are categorized as AML with MDS-related gene mutations and are associated with poor prognosis
- AML with myelodysplasia-related cytogenetic abnormalities
- This new category includes cases previously classified as AML with myelodysplasia-related changes (AML-MRC); the AML-MRC subgroup has been removed as genetic characteristics are now considered more relevant than clinical ones
- AML not otherwise specified (AML-NOS)
- This captures any other cases of AML, irrespective of the presence or absence of multilineage dysplasia
- Therapy-related AML is no longer considered a subtype, but it is a diagnostic qualifier to genetically-defined categories
- It is assumed to be a result of cytotoxic therapy-inducing mutations or the selection of minor clones resistant to chemotherapy
Figure 2. AML with recurrent genetic abnormalities*
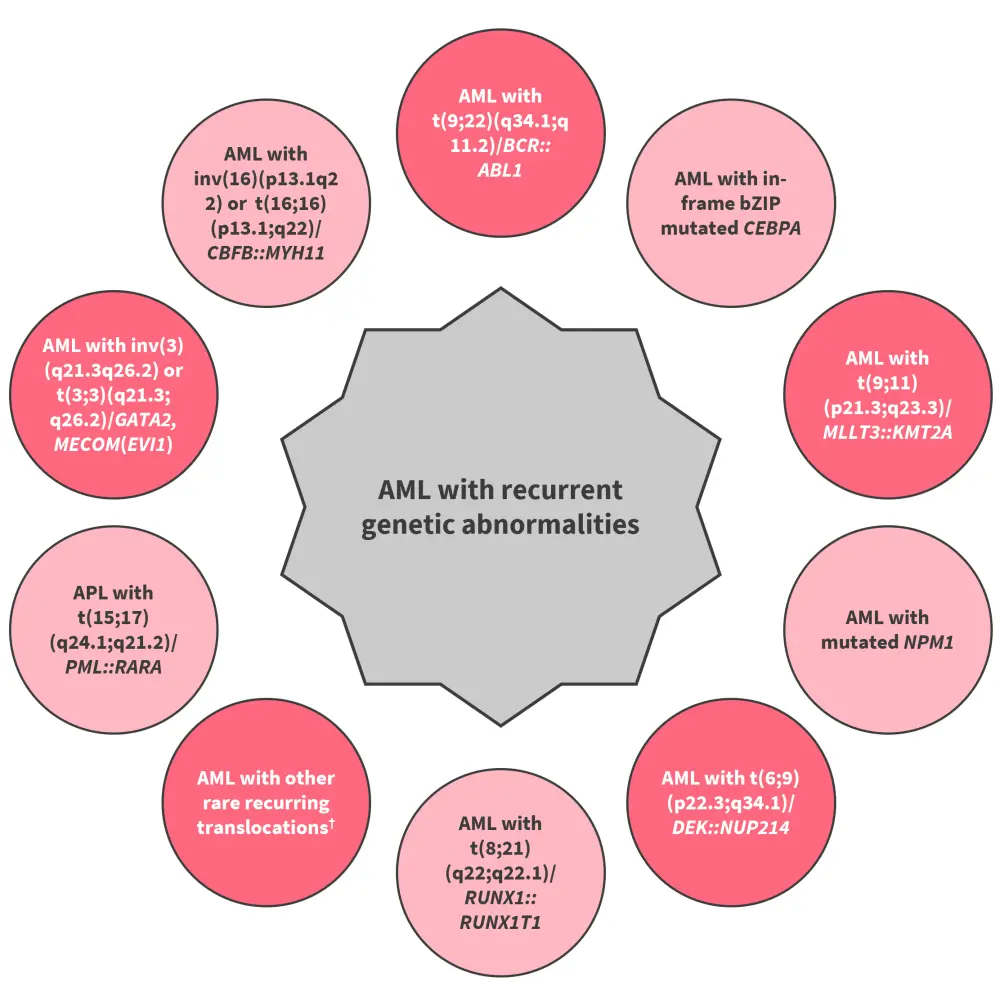
AML, acute myeloid leukemia; APL, acute promyelocytic leukemia.
*Data from Döhner, et al.1
†Other rare recurring translocations include AML with t(1;3)(p36.3;q21.3)/PRDM16::RPN1, AML (megakaryoblastic) with t(1;22)(p13.3;q13.1)/RBM15::MRTF1, AML with t(3;5)(q25.3;q35.1)/NPM1::MLF1, AML with t(5;11)(q35.2;p15.4)/NUP98::NSD1, AML with t(7;12)(q36.3;p13.2)/ETV6::MNX1, AML with t(8;16)(p11.2;p13.3)/KAT6A::CREBBP, AML with t(10;11)(p12.3;q14.2)/PICALM::MLLT10, AML with t(11;12)(p15.4;p13.3)/NUP98::KMD5A, AML with NUP98 and other partners. AML with t(16;21)(p11.2;q22.2)/FUS::ERG, AML with t(16;21)(q24.3;q22.1)/RUNX1::CBFA2T3, and AML with inv(16)(p13.3q24.3)/CBFA2T3::GLIS2
Germline predisposition
The risk of germline predisposition should be considered for all patients diagnosed regardless of age, given the impact on management, such as for allogeneic hematopoietic cell transplantation eligibility and to identify at-risk relatives. Clinicians should be familiar with testing options, including sequencing of skin fibroblasts. Only germline variants that are categorized as pathogenic or likely pathogenic are considered causative of disease and require follow-up within families.
A variant is deemed germline if:
- it is detected in DNA derived from a tissue source not likely to undergo somatic mutation frequently and at a variant allele frequency consistent with the germline (generally 30–60%); or
- it is identified in ≥2 relatives at a variant allele frequency consistent with the germline.
Testing for germline predisposition alleles is recommended when there is:
- a history of ≥2 cancers, one of which is a hematopoietic malignancy;
- a history of a hematopoietic malignancy plus another relative within two generations with a hematopoietic malignancy, a solid tumor diagnosed aged ≤50 years, or other hematopoietic abnormality;
- presence of a deleterious gene variant likely to be a germline allele (e.g., CHEK2 I200T and truncating DDX41 variants), especially if present during remission; or
- an earlier-than-average age of diagnosis (e.g., MDS diagnosed ≤40 years).
Diagnosis of AML
Immunophenotyping
Identification of cell surface and intracellular markers, such as CD13, CD33, and CD34, by immunophenotyping with multiparameter flow cytometry, is required to accurately diagnose AML. Immunohistochemistry on a core biopsy may be used if an aspirate is unable to be obtained.
Cytogenetic and molecular analyses
- Cytogenetic analysis is considered mandatory, with fluorescence in situ hybridization if required for some specific abnormalities.
- Molecular genetic testing should be used to screen for genetic abnormalities that define disease or risk categories, or those that are needed for targeted treatments.
- A dedicated gene panel for known predisposing alleles should be used when AML with germline predisposition is suspected.
- In NPM1-mutated and core-binding factor AML, quantitative polymerase chain reaction (PCR) or a droplet digital PCR is recommended to monitor measurable residual disease (MRD).
Biobanking
- BM and blood samples are recommended to be obtained at diagnosis, remission, and relapse, and stored under appropriate conditions with informed consent for laboratory studies.
The tests used to establish a diagnosis for a patient with AML are shown in Figure 3.
Figure 3. ELN recommended tests and procedures at diagnosis for patients with AML*
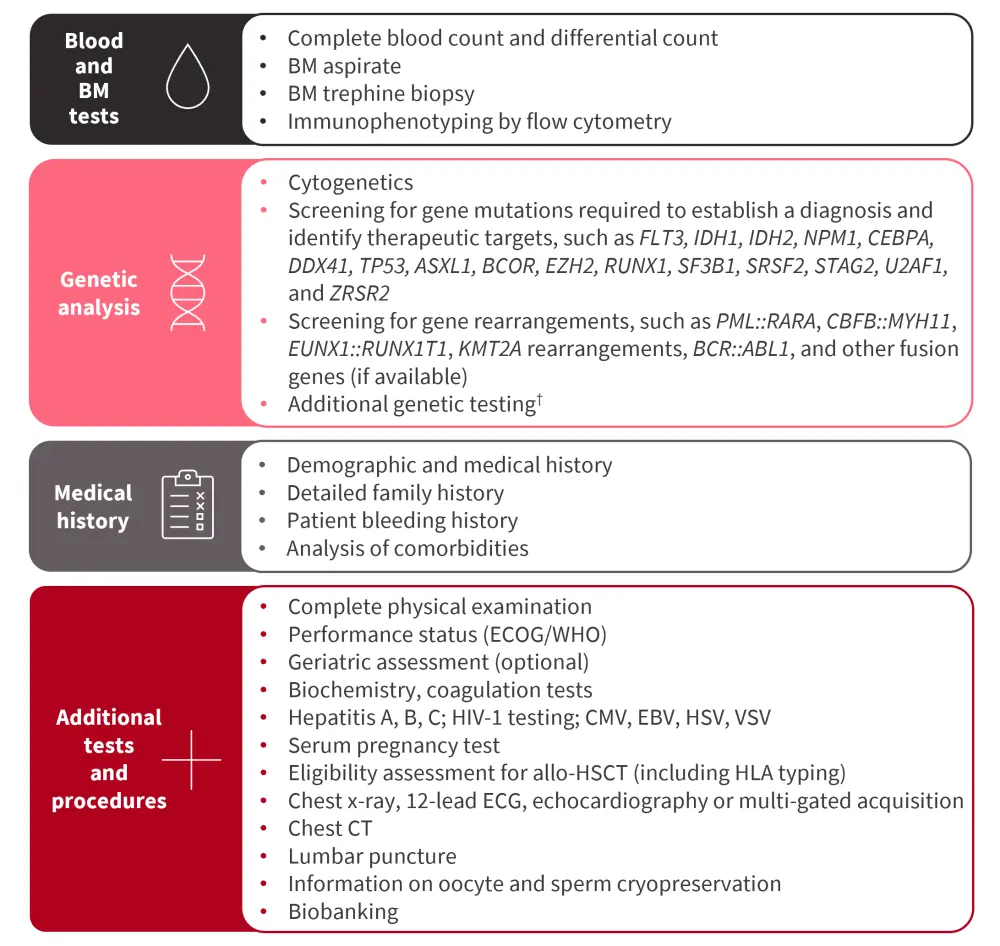
Allo-HSCT, allogeneic hematopoietic stem cell transplantation; AML, acute myeloid leukemia; BM, bone marrow; CT, computed tomography; CMV, cytomegalovirus; EBV, Epstein-Barr virus; ECG, electrocardiogram; ECOG, Eastern Cooperative Oncology Group; HLA, human leukocyte antigens; HSV, herpes simplex virus; VSV, varicella-zoster virus; WHO, World Health Organization.
*Adapted from Döhner, et al.1
†Additional genes recommended to test at diagnosis include ANKRD26, BCORL1, BRAF, CBL, CSF3R, DNMT3A, ETV6, GATA2, JAK2, KIT, KRAS, NRAS, NF1, PHF6, PPM1D, PTPN11, RAD21, SETBP1, TET2, and WT1.
2022 ELN risk classification at diagnosis
In the light of new data, response to initial therapy and early MRD testing are now considered to contribute to a patient’s risk classification, in addition to baseline genetic abnormalities (detailed in Table 1).
Changes from the previous recommendations include the following:
- All AML cases with FLT3-internal tandem duplication (ITD) should be classified as intermediate risk.
- AML with MDS-related gene mutations is classified as adverse risk.
- If adverse-risk cytogenetic abnormalities are present in NPM1-mutated AML, this indicates adverse risk.
- The presence of in-frame mutations affecting the bZIP region of CEBPA are classified as favorable risk.
- The adverse-risk group now includes recurring cytogenetic abnormalities, such as t(3q26.2;v) involving the MECOM gene or t(8;16)(p11;p13) associated with the KAT6A::CREBBP gene fusion.
- Hyperdiploid karyotypes with multiple trisomies are now considered adverse risk rather than complex karyotypes.
Table 1. 2022 ELN risk classification by genetics at initial diagnosis*
|
ELN, European LeukemiaNet. |
|
|
Risk category |
Genetic abnormality |
|---|---|
|
Favorable |
t(8;21)(q22;q22.1)/RUNX1::RUNX1T1 |
|
inv(16)(p13.1q22) or t(16;16)(p13.1;q22)/CBFB::MYH11 |
|
|
Mutated NPM1 without FLT3-ITD |
|
|
bZIP in-frame mutated CEBPA |
|
|
Intermediate |
Mutated NPM1 with FLT3-ITD |
|
Wild-type NPM1 with FLT3-ITD |
|
|
t(9;11)(p21.3;q23.3)/MLLT3::KMT2A |
|
|
Cytogenetic and/or molecular abnormalities not classified as favorable or adverse |
|
|
Adverse |
t(6;9)(p23;q34.1)/DEK::NUP214 |
|
t(v;11q23.3)/KMT2A-rearranged |
|
|
t(9;22)(q34.1;q11.2)/BCR::ABL1 |
|
|
t(8;16)(p11;p13)/KAT6A::CREBBP |
|
|
inv(3)(q21.3q26.2) or t(3;3)(q21.3;q26.2)/GATA2, MECOM(EVI1) |
|
|
t(3q26.2;v)/MECOM(EVI1)-rearranged |
|
|
−5 or del(5q); −7; −17/abn(17p) |
|
|
Complex karyotype, monosomal karyotype |
|
|
Mutated ASXL1, BCOR, EZH2, RUNX1, SF3B1, SRSF2, STAG2, U2AF1, or ZRSR2 |
|
|
Mutated TP53 |
|
Conclusion
These updated ELN 2022 recommendations regarding the diagnosis and classification of AML reflect the recent advancements in our understanding of AML, including the cytogenetic and mutational profiles involved.
References
Please indicate your level of agreement with the following statements:
The content was clear and easy to understand
The content addressed the learning objectives
The content was relevant to my practice
I will change my clinical practice as a result of this content


