All content on this site is intended for healthcare professionals only. By acknowledging this message and accessing the information on this website you are confirming that you are a Healthcare Professional. If you are a patient or carer, please visit Know AML.
The aml Hub website uses a third-party service provided by Google that dynamically translates web content. Translations are machine generated, so may not be an exact or complete translation, and the aml Hub cannot guarantee the accuracy of translated content. The aml and its employees will not be liable for any direct, indirect, or consequential damages (even if foreseeable) resulting from use of the Google Translate feature. For further support with Google Translate, visit Google Translate Help.
The AML Hub is an independent medical education platform, sponsored by Daiichi Sankyo, Johnson & Johnson, Syndax, Thermo Fisher Scientific, Kura Oncology, AbbVie, and has been supported through an educational grant from the Hippocrate Conference Institute, an association of the Servier Group.
The funders are allowed no direct influence on our content. The levels of sponsorship listed are reflective of the amount of funding given. View funders.
Now you can support HCPs in making informed decisions for their patients
Your contribution helps us continuously deliver expertly curated content to HCPs worldwide. You will also have the opportunity to make a content suggestion for consideration and receive updates on the impact contributions are making to our content.
Find out more
Create an account and access these new features:
Bookmark content to read later
Select your specific areas of interest
View AML content recommended for you
Measurable residual disease (MRD) is a readout used to inform prognosis and treatment decisions in patients with acute myeloid leukemia (AML), and there are currently two main techniques available to evaluate MRD, as shown in Table 1. An alternative approach is single-cell MRD analysis. During the 8th Annual Meeting of the Society of Hematologic Oncology (SOHO), Paresh Vyas, University of Oxford, UK, outlined the progress being made regarding single-cell MRD analysis in AML.1
Table 1. MRD analysis techniques1
|
LAIP, leukemia-associated immunophenotypes; LSC, leukemic stem cells; MRD, measurable residual disease; NGS, next-generation sequencing; NPM, nucleophosmin. |
|
|
Technique |
Sensitivity |
|---|---|
|
Molecular |
|
|
Single abnormality (NPM/fusion genes) |
10−4–10−6 |
|
NGS |
10−2 |
|
Flow cytometry |
10−3–10−4 |
|
LAIP flow |
|
|
LSC flow |
|
Introduction
On average, there are five recurrent driver mutations in adult de novo AML but, occurring separately, they do not all result in AML development. Paresh Vyas began by outlining the importance of understanding the clonal progression of hematopoietic cells (Figure 1) and the events that lead to AML. When considering therapy, it is necessary to understand which mutations occur in which cells, and which mutations occur in the same cell. This is because one therapy may prove lethal to cells with a particular mutational load while have no impact on alternative clones. Therefore, it becomes crucial to refine patient management based on the mutation combinations within a single cell.
Figure 1. Healthy hemopoiesis*

CMP, common myeloid progenitor; GMP, granulocyte-macrophage progenitor; LMPP, lymphoid-primed multipotent progenitor; MEP, megakaryocyte-erythrocyte progenitor; MPP, multipotent progenitor.
*Figure adapted from Vyas, 2020.1
Furthermore, not all clones are leukemic, with some residing in a preleukemic state that may never give rise to AML. Paresh Vyas defines preleukemic cells as those that are not arrested in differentiation, whereas leukemic cells are arrested in either the CD34+ progenitor or CD34− precursor stages (Figure 2). CD34+ AML comprises around 80% of human AML, and it has been shown that leukemic stem cells undergo arrest at the granulocyte-macrophage progenitor-like (GMP; CD38+) or the lymphoid-primed multipotent progenitor-like (LMPP; CD38−) stages. In a patient with AML, 20–90% of the bone marrow is made up of these leukemic clones with up to 107 times LMPP-like cell expansion.
Figure 2. Differentiation block in AML*
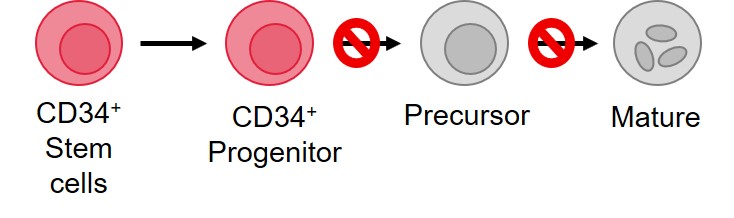
*Figure adapted from Vyas, 2020.1
Employing single-cell analysis
Paresh Vyas moved on to discuss how single-cell analysis assisted identification of the detailed mechanism of action (MoA) of enasidenib, an isocitrate dehydrogenase-2 (IDH2) mutant inhibitor. In patients with AML who achieved a complete response (CR) following treatment with single-agent enasidenib, the variant allelic frequency of IDH2 mutations was the same as prior treatment, indicating that enasidenib alleviates the differentiation block, shown in Figure 2. Flow-based analysis has provided further information on how enasidenib induces differentiation in these leukemic clones. Analysis of pre- and posttreatment cells showed a clear reduction in LMPP and GMP progenitor cells upon CR achievement. Combined, the findings show that not only does enasidenib induce maturation, but also a degree of restoration of stem cell compartment composition. In a patient example, single-cell analysis derived a complex cellular clonal structure, which Paresh Vyas suggested would have been ‘impossible to identify without single-cell analysis’. With this technology, the investigators were able to identify the exact mutations acquired during the leukemic clone evolution and the ones responsible for the later relapse of the patient. The findings from this group were imperative in understanding both the MoA of enasidenib and the mechanisms of relapse, outlining a number of pathways that could be therapeutically targeted to alleviate differentiation arrest.2 Importantly, these data highlight that defining the clonal structure could explain patient responses to therapy and the evolution of clones throughout treatment and relapse, as has been shown with gilteritinib. Clonal responses to gilteritinib, both during treatment and following relapse, have been identified with the help of single-cell mutation analysis, uncovering the precise AML clonal landscape.3
Conclusion
Platforms combining single-cell mutational analysis with transcriptome analysis are becoming increasingly available. These emerging technologies will help to greater define the impact of treatment on leukemic clonal structure, on the epigenome and the transcriptome, as well as the mechanisms behind AML resistance.
References
Please indicate your level of agreement with the following statements:
The content was clear and easy to understand
The content addressed the learning objectives
The content was relevant to my practice
I will change my clinical practice as a result of this content


 Paresh Vyas
Paresh Vyas