All content on this site is intended for healthcare professionals only. By acknowledging this message and accessing the information on this website you are confirming that you are a Healthcare Professional. If you are a patient or carer, please visit Know AML.
The aml Hub website uses a third-party service provided by Google that dynamically translates web content. Translations are machine generated, so may not be an exact or complete translation, and the aml Hub cannot guarantee the accuracy of translated content. The aml and its employees will not be liable for any direct, indirect, or consequential damages (even if foreseeable) resulting from use of the Google Translate feature. For further support with Google Translate, visit Google Translate Help.
The AML Hub is an independent medical education platform, sponsored by Daiichi Sankyo, Johnson & Johnson, Syndax, Thermo Fisher Scientific, Kura Oncology, AbbVie, and has been supported through an educational grant from the Hippocrate Conference Institute, an association of the Servier Group.
Funders are allowed no direct influence on our content. The levels of sponsorship listed are reflective of the amount of funding given. View funders.
Now you can support HCPs in making informed decisions for their patients
Your contribution helps us continuously deliver expertly curated content to HCPs worldwide. You will also have the opportunity to make a content suggestion for consideration and receive updates on the impact contributions are making to our content.
Find out more
Create an account and access these new features:
Bookmark content to read later
Select your specific areas of interest
View AML content recommended for you
Standardizing next-generation sequencing MRD assessment in AML
Detection of measurable residual disease (MRD) is recommended for the evaluation of treatment response in patients with acute myeloid leukemia (AML). Variation in methods and lack of standardization limits the comparability of MRD assessments between different laboratories. Next-generation sequencing (NGS) is a promising method for MRD analysis, but reproducibility across laboratories has not been evaluated.1
During the European Hematology Association (EHA) 2021 Virtual Congress, Michael Heuser presented data from an international study by the ELN AML Working Party evaluating the reproducibility of NGS MRD analysis across laboratories.1 Here, we are pleased to provide a summary of the results.
Study design
A total of 13 laboratories across Europe and the US participated in the study. Both the local NGS MRD approach of the participating laboratory (local protocol) and a standardized central NGS MRD approach (central protocol) were assessed. The study design is shown in Figure 1.
Figure 1. Study design*
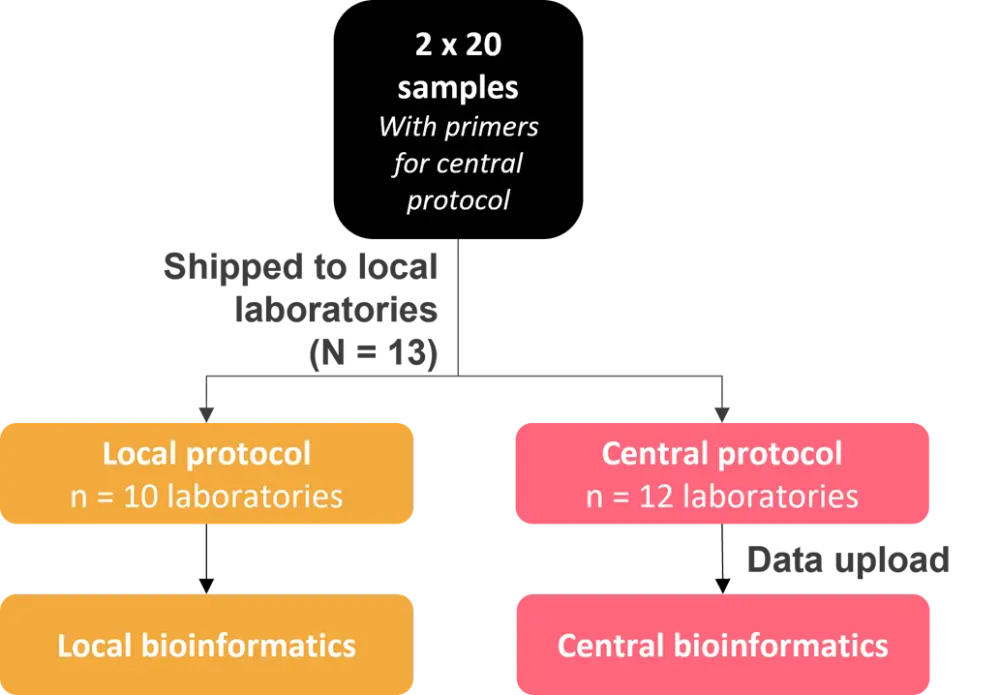
*Figure adapted from M. Heuser.1
The following six gene mutations were analyzed:
- IDH1 R132C
- IDH1 R132H
- IDH2 R140Q
- KIT D816V
- NPM1 type A
- TP53 R248Q
A total of 24 variants were present in 18 MRD-positive samples. Two samples were MRD negative, and these were assessed for IDH2 and KIT mutations. Variant allele frequencies (VAF) ranged from 1.55% to 0.004%.
Local sequencing was performed by either error-corrected targeted amplicon sequencing or myeloid panel sequencing with/without error correction. For the central MRD approach, targeted amplicon sequencing of known targets was performed with error correction using genetic barcodes.
Study results
Sequencing reads
Using local protocols, the number of aligned sequencing reads was often similar within each laboratory, but different among the participating laboratories. In contrast, when using the central protocol, there were similar results within each laboratory and between different laboratories.
Limit of detection
The VAF limit of detection (LOD) ranged from 0.003% to 1% for the local protocols. For the central protocol, the LOD ranged from 0.001% to 1%, and was similar within laboratories and among different laboratories.
Repeatability
To assess repeatability of VAF measurements within individual laboratories, six variants were quantified in duplicate and the coefficient of variation (CoV) was calculated for each duplicate by each laboratory. There was good repeatability for most local protocols, with a median CoV of 13%. Repeatability was further improved using the central protocol, with a median CoV of just 6.3%.
Linearity
Linearity was assessed using the IDH1 R132H mutation, which had a VAF of 1.2% in the undiluted sample. The 8-fold dilution was linearly quantified for all local and central protocols. A further 4-fold dilution remained linear for some local protocols and for the central protocol. Linearity over a large concentration range was also confirmed across all protocols for the NPM1 mutation.
Reproducibility
For local protocols, reproducibility of VAF measurements between laboratories was very good for 1% VAFs but declined at lower VAFs; whereas, for the central protocol, reproducibility was very good for VAFs as low as 0.01%.
The LOD for local protocols, at around 0.1%, may explain the poor reproducibility seen for lower VAFs. In contrast, the LOD for the central protocol was around 0.01%, and the variants could therefore be clearly distinguished from background noise. These low-level variants would be defined as negative according to the 0.1% recommended cutoff for NGS-MRD, causing some patients with detectable MRD to be misclassified as MRD negative. The prognostic implication of this low-level MRD, however, remains to be determined.
Sensitivity and specificity
The sensitivity and specificity for detecting MRD positive (n = 24) and negative (n = 2) samples using local protocols was 100% for 6/10 laboratories and 8/10 laboratories, respectively. All 12 laboratories using the central protocol correctly identified the MRD-positive and -negative samples (100% sensitivity and specificity).
Conclusion
This first interlaboratory analysis of NGS MRD in AML showed that local NGS MRD methods were highly sensitive and specific for most participating laboratories, and a standardized, highly reproducible protocol for NGS MRD could be successfully incorporated into local laboratories.
References
Please indicate your level of agreement with the following statements:
The content was clear and easy to understand
The content addressed the learning objectives
The content was relevant to my practice
I will change my clinical practice as a result of this content

